Gaussian Process Emulator with Functional Data#
In many scenarios, we may be dealing with functional data. This means that the input, output, or both, are sampled over a particular dimension (e.g. time). In twinlab, data are presented in column-feature format, meaning a single data sample of functional format may contain hundreds or thousands of columns.
Gaussian Process models do not scale well to these scenarios, so we provide the ability to perform dimensionality reduction before model fitting.
This notebook will cover:
How to decompose functional inputs and outputs
[ ]:
# Third party imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from itertools import product
# twinLab import
import twinlab as tl
Problem Formulation#
Here, we define a problem with two input dimensions and one functional output defined over a grid of sample locations
[ ]:
# Grid over which the functional output is defined
grid = np.linspace(0, 1)
# True function: Forrester function with variable a and b
def model(x):
return (x[0] * grid - 2) ** 2 * np.sin(x[1] * grid - 4)
# Define input data
x = np.random.uniform(size=(100, 2))
x[:, 0] = x[:, 0] * 4 + 4
x[:, 1] = x[:, 1] * 4 + 10
# Compute output data
y = np.zeros((x.shape[0], grid.size))
for i, x_i in enumerate(x):
y[i, :] = model(x_i)
y = {"y_{}".format(i): y[:, i] for i in range(grid.size)}
# Save to DataFrame
df = pd.DataFrame({"x1": x[:, 0], "x2": x[:, 1], **y})
df.head()
[ ]:
# Define the name of the dataset
dataset_id = "FunctionalGP_Data"
# Intialise a Dataset object
dataset = tl.Dataset(id=dataset_id)
# Upload the dataset
dataset.upload(df, verbose=True)
Functional Emulator Workflow#
In twinlab, dimensionality reduction is implemented in the form of truncated Singular Value Decomposition (tSVD), and is accessible in two ways. It can be performed by specifiying the number of dimensions we want to truncate the data to using input_retained_dimensions for inputs and output_retained_dimensions for outputs. This can also be specified through the amount of variance to be explained by the data through input_explained_variance for inputs and
output_explained_variance for outputs. These parameters are part of the TrainParams object, and can then be further passed to the training function.
One can decompose the inputs, outputs, or both in the same Emulator.
[ ]:
# Initialise emulator
emulator_id = "FunctionalGP"
emulator = tl.Emulator(id=emulator_id)
# Define the training parameters for your emulator
output_columns = df.columns[2:].to_list()
params = tl.TrainParams(
train_test_ratio=0.75,
estimator="gaussian_process_regression",
input_retained_dimensions=1,
output_explained_variance=0.99,
)
# Train the emulator using the train method
emulator.train(
dataset=dataset,
inputs=["x1", "x2"],
outputs=output_columns,
params=params,
verbose=True,
)
[5]:
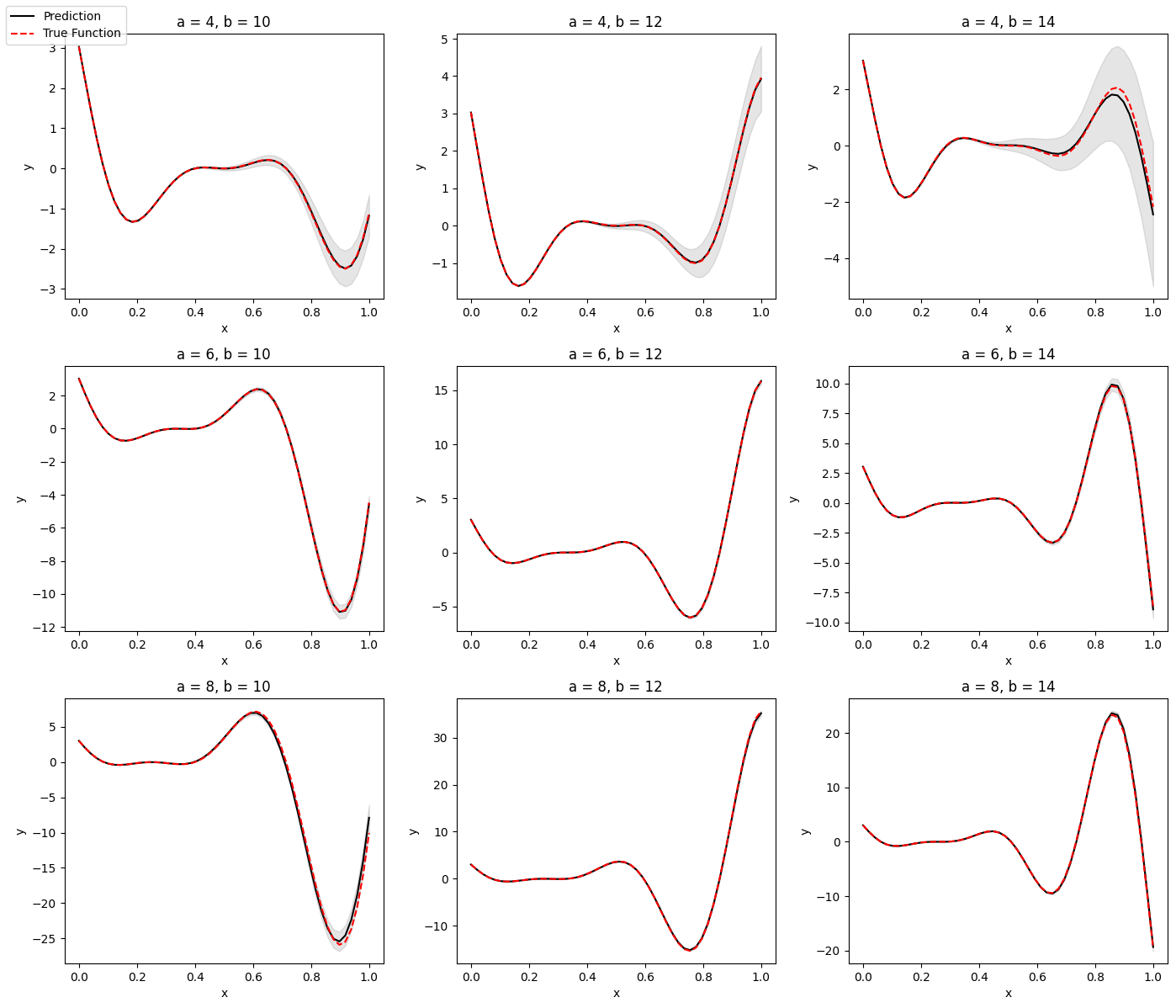
# Create grid of output plots
grid = np.linspace(0, 1)
x1 = [4, 6, 8]
x2 = [10, 12, 14]
X = np.array(list(product(x1, x2)))
ax_i = list(product([0, 1, 2], [0, 1, 2]))
# Create output plot and save in directory
fig, axes = plt.subplots(figsize=(14, 12), nrows=3, ncols=3)
# Setup legend
legend_labels = {}
for i, x_i in enumerate(X):
r, c = ax_i[i]
ax = axes[r, c]
X_test = pd.DataFrame(x_i[np.newaxis, :], columns=["x1", "x2"])
y_test = model(X_test.values.flatten())
y_pred = emulator.predict(X_test)
y_df = pd.concat([y_pred[0], y_pred[1]], axis=0)
y_mean, y_stdev = y_df.iloc[0, :], y_df.iloc[1, :]
y_mean = y_mean.values
y_stdev = y_stdev.values
ax.set_title("a = {}, b = {}".format(x_i[0], x_i[1]))
ax.plot(grid, y_mean.flatten(), c="k")
ax.plot(grid, y_test.flatten(), c="red", linestyle="dashed")
ax.fill_between(
grid,
(y_mean - 1.96 * y_stdev).flatten(),
(y_mean + 1.96 * y_stdev).flatten(),
color="k",
alpha=0.1,
)
ax.set_xlabel("x")
ax.set_ylabel("y")
# Store labels for the legend
legend_labels[f"Prediction"] = plt.Line2D([0], [0], color="k")
legend_labels[f"True Function"] = plt.Line2D([0], [0], color="red", linestyle="dashed")
# Print legend
fig.legend(legend_labels.values(), legend_labels.keys(), loc="upper left")
fig.tight_layout()
[6]:
# Delete emulator and dataset
emulator.delete()
dataset.delete()
